
Geir Evensen, Chief Scientist at NORCE Norwegian Research Centre AS
Data assimilation is key to my COVID-19 modelling work, and I was pleased to be invited to talk about it in more detail in an ECMWF blog and seminar (to be live-streamed on 30 September). The onset of the SARS-COV-2 (or COVID-19) pandemic led to a complete lockdown of society in many countries. So there I was, an applied mathematician, in confinement, following the news coverage of extreme situations in hospitals in Italy and New York, and at the same time hearing confusing and contradictory statements from politicians and leaders in various countries.
This situation motivated me to contribute with knowledge-based information to support decision-makers. So, in my home office, I started developing an epidemic model and gathering data on the COVID-19 related hospitalisations and deaths in Norway. I coupled the model to my data-assimilation library, and at the end of March, after about two weeks of work, I was able to model and predict the SARS-COV-2 pandemic evolution in Norway. I used the data-assimilation system to calibrate model parameters, including the time-dependent effective-reproductive-number, R. I then provided reports with model predictions to the Norwegian authorities to explain the unstable situation and the pandemic's dependency on R.
On 10 April, I invited an international group of colleagues working in data assimilation to use the model system on data from their respective countries. Nearly everyone jumped at the opportunity, and from then, we have worked together as an international team. We modelled the SARS-COV-2 pandemic in four European countries, Norway, England, the Netherlands, and France; the province of Quebec in Canada; the South American countries Argentina and Brazil; and the four US states Alabama, North Carolina, California, and New York. These countries and states all have vastly different developments of the epidemic, and we could accurately model the SARS-CoV-2 outbreak in all of them.
The joint work led to significant learnings regarding the use of data assimilation in epidemic modelling. We compiled the results into a large manuscript and submitted it to the journal "Fundamentals of Data Science" and we are currently finalising the revisions. We also made the paper available from MedRxiv.
We used a SEIR (susceptible, exposed, infectious, recovered) model with age-classes and compartments of sick, hospitalised, and deceased (Figure 1).
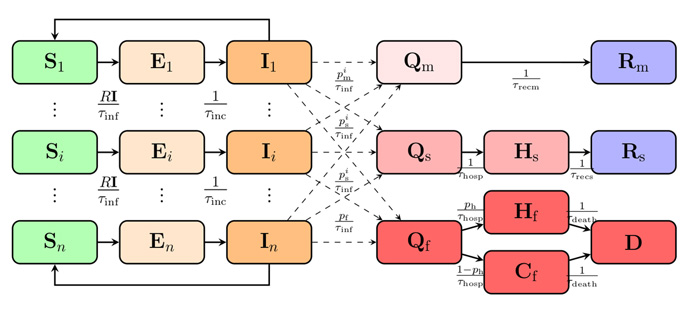
Figure 1: A schematic of the SEIR model. Populations of susceptible, exposed and infectious are divided into age groups Si, Ei, and Ii. S = susceptible, E = exposed, I = infectious, R = recovered, Q = quarantined, H = hospitalised, C = home/care home, D = deceased. The different age groups of infectious Ii , transition into the different quarantined groups of sick based on the fractions p i m , p i s , p i f , which refer to the portion of patients with mild symptoms, the hospitalised fraction of patients with severe symptoms, and the fraction of fatally ill.
This work demonstrates how it is possible to use iterative ensemble smoothers to estimate parameters in a SEIR model. The data assimilated are the daily numbers of accumulated deaths and the number of hospitalised. Also, it is possible to condition the model on the number of cases obtained from testing.
We start from a wide prior distribution representing the model parameters; then, the ensemble conditioning leads to a posterior ensemble of estimated parameters leading to model predictions in close agreement with the observations (see examples in Figure 2). The updated ensemble of model simulations has predictive capabilities and includes uncertainty estimates.
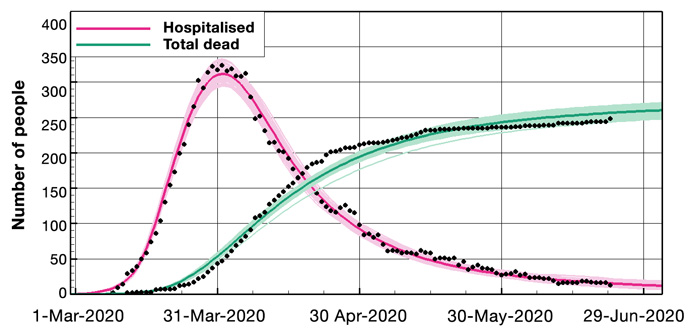
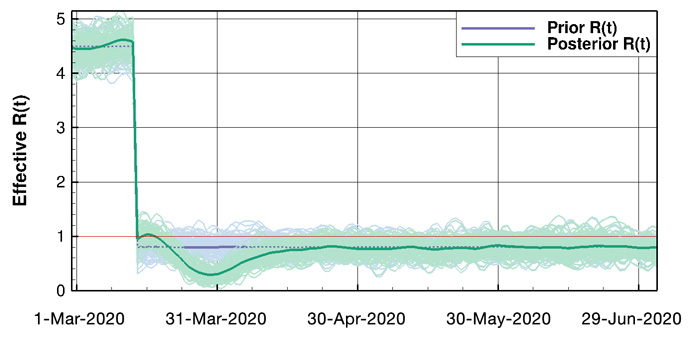
Figure 2: Example results for Norway. Above: Ensemble means and the 100 first ensemble realisations, for the number of hospitalised and the accumulated amount of deaths. Below: The prior and posterior ensembles of R(t).
We estimate the effective-reproductive-number (R) as a function of time, and we can assess the impact of different intervention measures. By starting from the updated set of model parameters, we can make accurate short-term predictions of the epidemic development given knowledge of the future effective-reproductive-number. Also, the model system allows for the computation of long-term scenarios of the epidemic under different assumptions.
The model system is freely available from Github.
We realise that more complex models, e.g. with regional compartments, may be desirable, and we suggest that the approach used here should be applicable also for these models. I have recently upgraded the model system to include multiple regions or countries with specified interactions between them. This new model formulation allows for simulating the pandemic development for, for example, the countries in Europe or the states in the US.
Seminar on 30 September
Geir will present this work in a virtual seminar at ECMWF on 30 September 2020, 14:15 BST. The seminar will be live-streamed and is open to all.